
Adapter and primer sequences: Next GEM Single Cell 5 Gel Beads v2 (PN. Join us for this webinar to hear about our latest advances in Targeted Gene Expression, Chromium Single Cell Immune Profiling v2 with Feature Barcode Technology, and the Visium Spatial Gene Expression Solution with Immunofluorescence. Im not an expert on this, so Im not able to comment on the details of these primers. At 10x Genomics, we provide single cell and spatial solutions that enable you to drive the leading edge of what’s possible.
Speaker: Science & Technology Advisor, Leo Chan, Ph.D.,Īs we enter a century where transformative advances will reshape the way we examine biological complexities from single cells and tissues,researchers need approaches that can keep up with the fast pace of their science. Title: Latest Advances in 10x Genomics’ Single Cell and VisiumSpatial Gene Expression Solutions. This will be a precious opportunity for you to learn the latest advances in Targeted Gene Expression, Chromium Single Cell Immune Profiling v2 with Feature Barcode Technology, and the Visium Spatial Gene Expression Solution with Immunofluorescence. He will introduce Latest Advances in 10x Genomics. The pipeline also recomputes the V(D)J clonotype grouping on the combined data.DNA sequencing section would like to invite you to the following seminar by Dr.Leo Chan. The aggr pipeline normalizes the individual gene expression and feature barcode runs to the same sequencing depth, recomputes the feature-barcode matrices and performs analysis on the combined data. Additionally, the cell calls provided by the gene expression data are used to improve the cell calls inferred by the V(D)J library.Ĭellranger aggr aggregates outputs from multiple runs of cellranger count/vdj/multi. It also performs sequence assembly and paired clonotype calling on the V(D)J libraries. It performs alignment, filtering, barcode counting, and UMI counting on the Gene Expression and/or Feature Barcode libraries. cloupe file which can be loaded into Loupe Browser for interactive visualization, clustering, and differential expression analysis.Ĭellranger multi takes FASTQ files from cellranger mkfastq or bcl2fastq for any combination of 5' Gene Expression, Feature Barcode (cell surface protein or antigen) and V(D)J libraries from a single gem-well. It uses the Chromium cellular barcodes to generate feature-barcode matrices, determine clusters, and perform gene expression analysis. vloupe file which can be loaded into Loupe V(D)J Browser.Ĭellranger count takes FASTQ files from cellranger mkfastq or bcl2fastq for 5' Gene Expression and/or Feature Barcode (cell surface protein or antigen) libraries and performs alignment, filtering, barcode counting, and UMI counting. Clonotypes and CDR3 sequences are output as a. It uses the Chromium cellular barcodes and UMIs to assemble V(D)J transcripts per cell. It is a wrapper around Illumina's bcl2fastq, with additional useful features that are specific to 10x libraries and a simplified sample sheet format.Ĭellranger vdj takes FASTQ files from cellranger mkfastq or bcl2fastq for V(D)J libraries and performs sequence assembly and paired clonotype calling. Please check the core ( or 71) for the detail information.Ĭellranger mkfastq demultiplexes raw base call (BCL) files generated by Illumina sequencers into FASTQ files. The nuclei will be checked the quality and the core has the rights to determine if the samples are qualified for the experiment. 
Samples have to be sent to core before 1 PM.
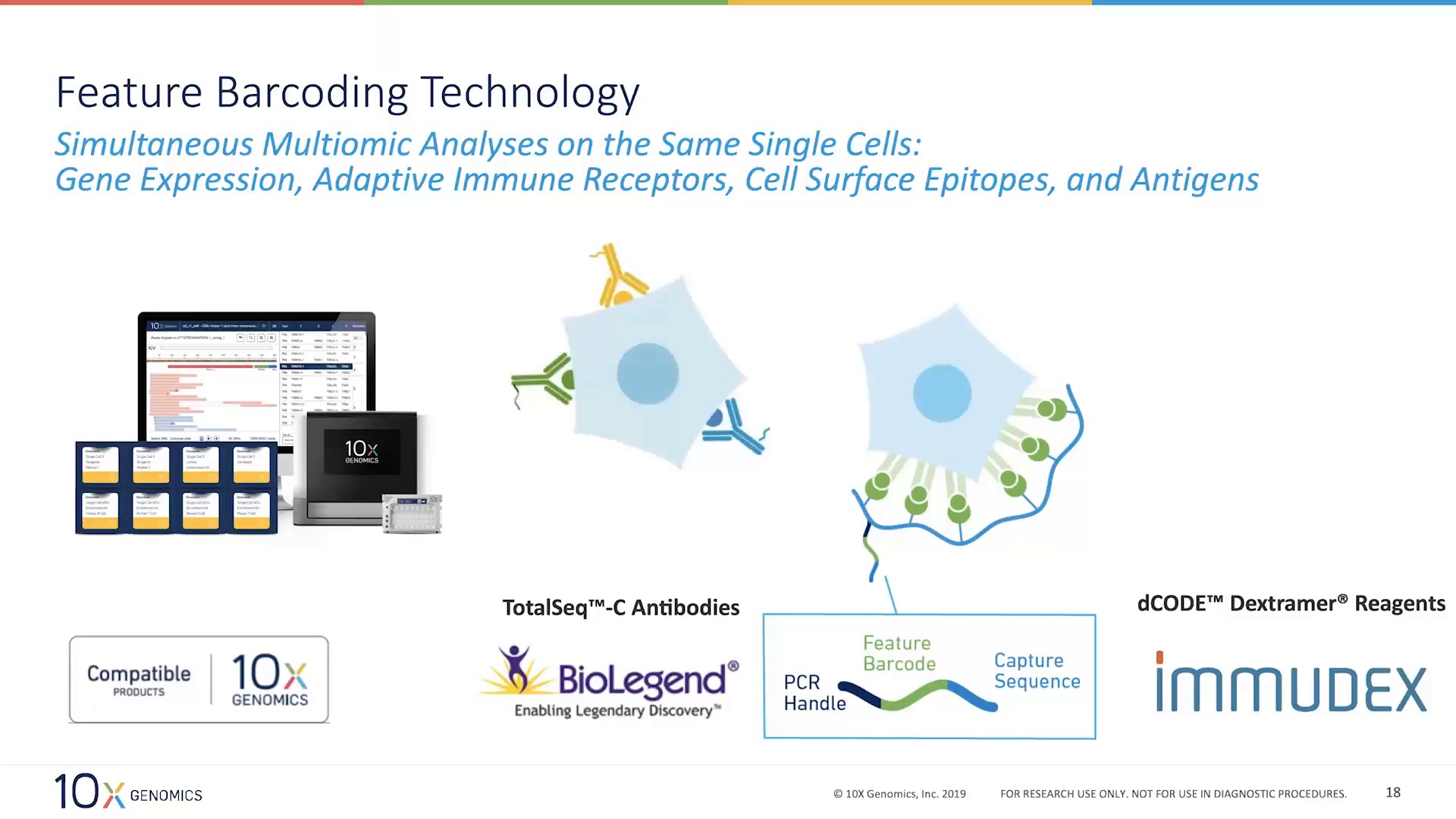
Single cells are suspended in 1xPBS with 0.04%BSA.Characterize immune response to infection by measuring clonal expansion and immune cell phenotypes.Perform large-scale antibody and TCR discovery for novel antigens.Analyze tissue microenvironment, disease progression, and drug immune response.Identify and characterize rare cell types and biomarkers.Explore adaptive and innate immune cell diversity.This service is able to 1) Profile thousands of genes at the single cell level by barcoding mRNA at the 5’ end, for unbiased characterization of cell types and cell states 2) Simultaneously profile immune repertoire (BCR/TCR) and gene expression from the same cell to enable correlation of clonotype with the corresponding cell subtype 3) Obtain paired, full-length receptor sequences from T cells and/or B cells with complete isotype resolution, providing functional data for antibody/TCR discovery. Discover new cell types and states using whole transcriptome analysis, or focus your search with targeted gene expression panels of interest. ScRNA Immune (Single-cell RNA Immune profiling, 10x Genomics 5’ kit)Ĭhromium Single Cell Immune Profiling provides a comprehensive approach to simultaneously examine cellular heterogeneity of the immune system, T- and B-cell repertoire diversity, and antigen specificity at single cell resolution.







 0 kommentar(er)
0 kommentar(er)
